Simulating the Distance Distribution between Spin-Labels Attached to Proteins
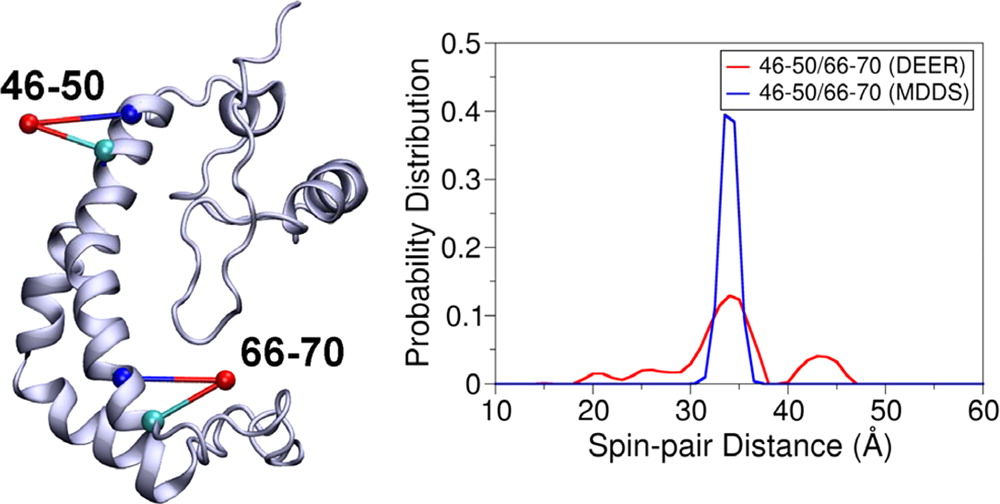

Structural Dynamics of Protein Interactions Using Site-Directed Spin Labeling of Cysteines to Measure Distances and Rotational Dynamics with EPR Spectroscopy

Spin Label - an overview

Spin Label - an overview
DEER data of spin-labeled Bax WT variants in solution and in membranes.
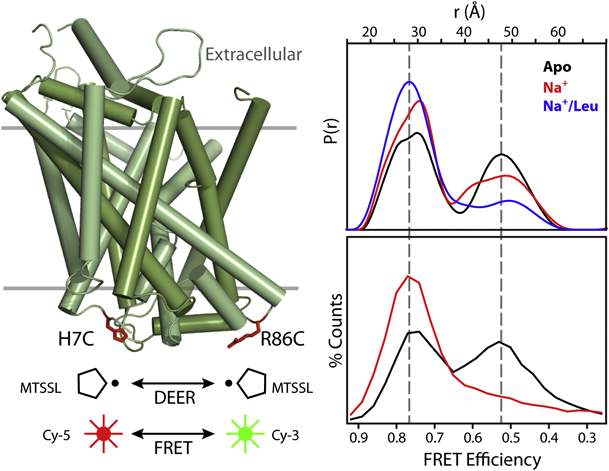
Toward the fourth dimension of membrane protein structure: Insight into dynamics from spin-labeling EPR spectroscopy

Nanoscale lipid membrane mimetics in spin-labeling and electron paramagnetic resonance spectroscopy studies of protein structure and function

Site-directed spin labeling-electron paramagnetic resonance spectroscopy in biocatalysis: Enzyme orientation and dynamics in nanoscale confinement - ScienceDirect

Molecular Dynamics Simulations Based on Newly Developed Force Field Parameters for Cu2+ Spin Labels Provide Insights into Double-Histidine-Based Double Electron–Electron Resonance

Frontiers Characterization of Weak Protein Domain Structure by Spin-Label Distance Distributions
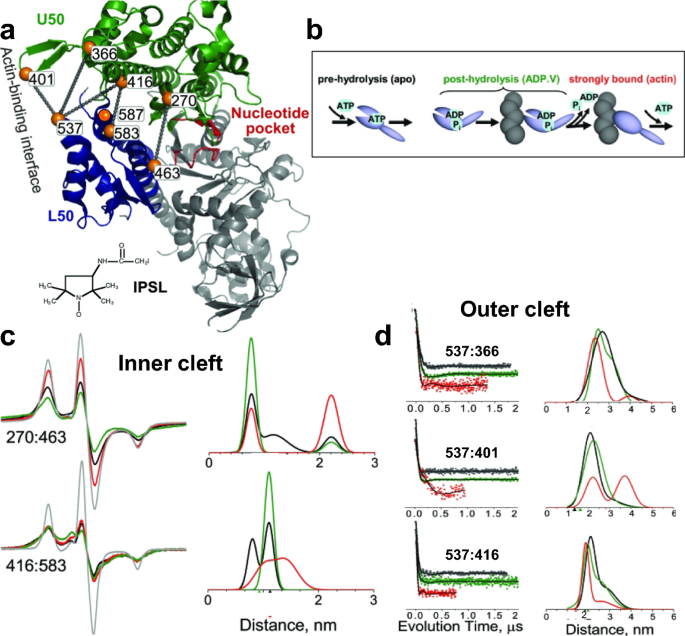
Structural Dynamics of Protein Interactions Using Site-Directed Spin Labeling of Cysteines to Measure Distances and Rotational Dynamics with EPR Spectroscopy

Structural Dynamics of Protein Interactions Using Site-Directed Spin Labeling of Cysteines to Measure Distances and Rotational Dynamics with EPR Spectroscopy

Simulating the Distance Distribution between Spin-Labels Attached to Proteins
Spin Labels in Electron Spin Resonance (ESR) Spectroscopy

Figure 2 from DEER EPR measurements for membrane protein structures via bifunctional spin labels and lipodisq nanoparticles.